Intro to R in Epidemiology – RMarkdown Edition!
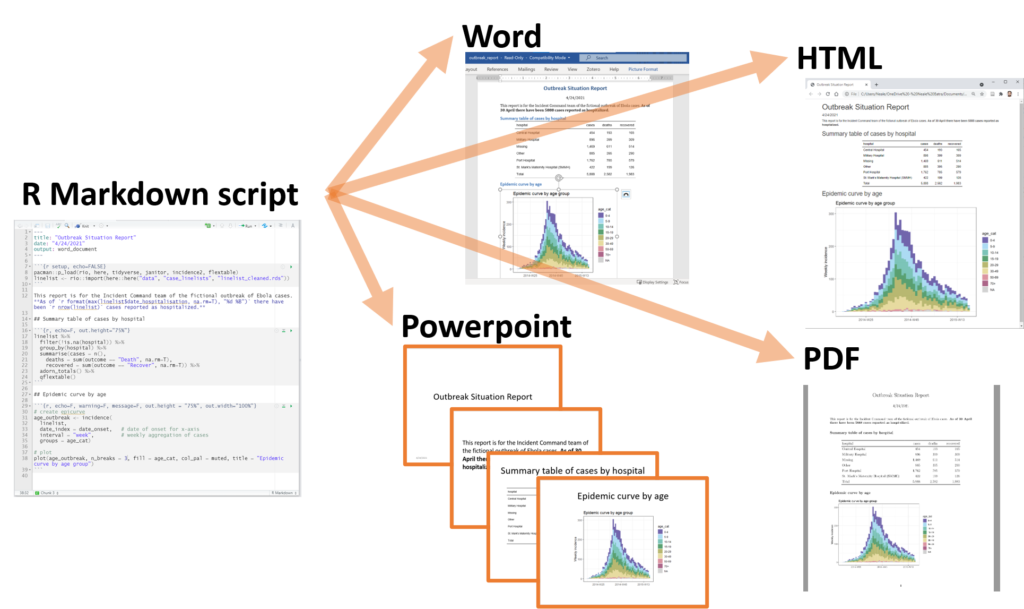
Taken from https://www.epirhandbook.com/new_pages/rmarkdown.html
R Markdown is a widely-used tool for creating automated, reproducible, and share-worthy outputs, such as reports. It can generate static or interactive outputs, in Word, pdf, html, powerpoint, and other formats.
An R Markdown script intersperces R code and text such that the script actually becomes your output document. You can create an entire formatted document, including narrative text (can be dynamic to change based on your data), tables, figures, bullets/numbers, bibliographies, etc.
Such documents can be produced to update on a routine basis (e.g. daily surveillance reports) and/or run on subsets of data (e.g. reports for each jurisdiction).
Background to R Markdown
To explain some of the concepts and packages involved:
- Markdown is a “language” that allows you to write a document using plain text, that can be converted to html and other formats. It is not specific to R. Files written in Markdown have a ‘.md’ extension.
- R Markdown: is a variation on markdown that is specific to R – it allows you to write a document using markdown to produce text and to embed R code and display their outputs. R Markdown files have ‘.Rmd’ extension.
- rmarkdown – the package: This is used by R to render the .Rmd file into the desired output. It’s focus is converting the markdown (text) syntax, so we also need…
- knitr: This R package will read the code chunks, execute it, and ‘knit’ it back into the document. This is how tables and graphs are included alongside the text.
- Pandoc: Finally, pandoc actually convert the output into word/pdf/powerpoint etc. It is a software separate from R but is installed automatically with RStudio.
In sum, the process that happens in the background (you do not need to know all these steps!) involves feeding the .Rmd file to knitr, which executes the R code chunks and creates a new .md (markdown) file which includes the R code and its rendered output. The .md file is then processed by pandoc to create the finished product: a Microsoft Word document, HTML file, powerpoint document, pdf, etc.
Installation
To create a R Markdown output, you need to have the following installed:
- The rmarkdown package (knitr will also be installed automatically)
- Pandoc, which should come installed with RStudio. If you are not using RStudio, you can download Pandoc here: http://pandoc.org.
pacman::p_load(rmarkdown)
Starting a new Rmd file
In RStudio, open a new R markdown file, starting with ‘File’, then ‘New file’ then ‘R markdown…’. R Studio will give you some output options to pick from. You can select HTML for simplicity. The title and the author names are not important. If the output document type you want is not one of these, don’t worry – you can just pick any one and change it in the script later.


Responses