Intro to R in Epidemiology (Part Two)
Written by Aashna Uppal @auppal
(The content of this course draws from the EpiR Handbook (Chapters 3-8, 17, 30, 40), which is a free online resource developed by Applied Epi)
- Base R vs. tidyverse
- The tidyverse is like a package of packages, designed specifically for working with data in a “tidy” way.
- You can still manipulate data without it – this is called the “base R” way of doing it.
- The difference between the two can be illustrated by a quick example.
- In order to bake a cake, you need to
put_in_oven(mix_all_ingredients_together_into_a_cake_batter(combine(flour, egg, baking_soda, butter, sugar)))
- In order to bake a cake, you need to
flour %>%
add(egg) %>%
add(baking_soda) %>%
add(butter) %>%
add(sugar) %>%
mix_all_ingredients_together_into_a_cake_batter() %>%
put_in_oven()The pipe operater (%>%)
- Simply explained, the pipe operator (
%>%) passes an intermediate output from one function to the next. You can think of it as saying “then”. Many functions can be linked together with %>%. - Piping emphasizes a sequence of actions, not the object the actions are being performed on
- Pipes come from the package magrittr, which is automatically included in packages dplyr and tidyverse
- Pipes can make code more clean and easier to read, more intuitive
- Importing data
- When you import a “dataset” into R, you are generally creating a new data frame object in your R environment and defining it as an imported file (e.g. Excel, CSV, TSV, RDS) that is located in your folder directories at a certain file path/address. We will see an example of this later.
- You can import/export many types of files, including those created by other statistical programs (SAS, STATA, SPSS). You can also connect to relational databases.
The rio package
- The R package we recommend for importing data is
rio. - Its functions
import()andexport()can handle many different file types (e.g. .xlsx, .csv, .rds, .tsv). When you provide a file path to either of these functions (including the file extension like “.csv”),riowill read the extension and use the correct tool to import or export the file. - The alternative to using
riois to use functions from many other packages, each of which is specific to a type of file. For example,read.csv()(base R),read.xlsx()(openxlsx package), andwrite_csv()(readr pacakge), etc.
The here package
- The package here and its function
here()make it easy to tell R where to find and to save your files – in essence, it builds file paths. - Used in conjunction with an R project,
hereallows you to describe the location of files in your R project in relation to the R project’s root directory (the top-level folder). This is useful when the R project may be shared or accessed by multiple people/computers. - When the here package is first loaded within the R project, it places a small file called “.here” in the root folder of your R project as a “benchmark” or “anchor”
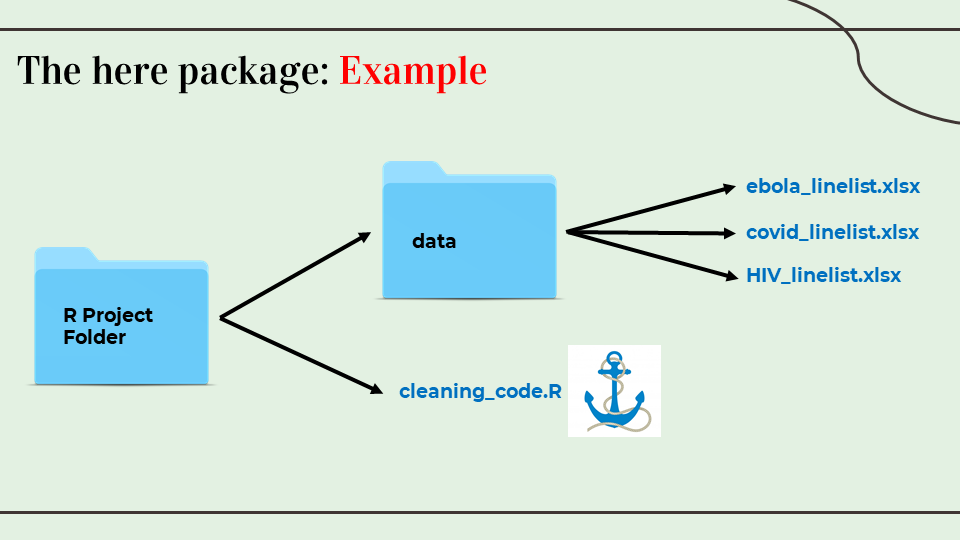
here::i_am("cleaning_code.R")
linelist <- import(here("data", "ebola_linelist.xlsx"))Written by Aashna Uppal @auppal



Responses