Hell no, fitting 16 regressions at the same time.
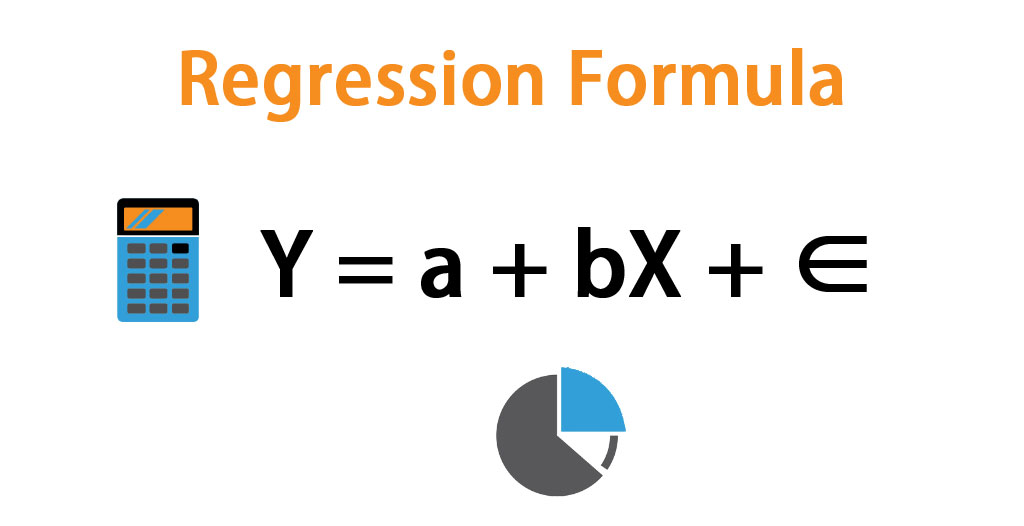
Straight into the pool
This post assumes you are already familiar with regression. No attempt will be made to explain the concept or justify any decision. The purpose of this post is to demonstrate a cool feature you can use to fit several models at once.
Required packages
- tidyverse (of course, 😀)
- wakefield (to generate random data)
- gtsummary (for table outputs)
Problem
For a strange reason, which I recently encountered, I had to fit a different regression for each region in the country to test the true effect of what i was seeing.
I could fit it one by one, a pain it would be. But then, the only reason why we write computer programs is because we are lazy and smart too. We use our smartness to benefit ourselves.
require(wakefield)Loading required package: wakefieldrequire(ggpubr)Loading required package: ggpubrLoading required package: ggplot2require(gtsummary)Loading required package: gtsummary#StandWithUkrainerequire(tidyverse) ## If you are an ardent follower of tidyverse, always load it lastLoading required package: tidyverse── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ lubridate 1.9.3 ✔ tibble 3.2.1
✔ purrr 1.0.2 ✔ tidyr 1.3.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ lubridate::hour() masks wakefield::hour()
✖ dplyr::id() masks wakefield::id()
✖ lubridate::interval() masks wakefield::interval()
✖ dplyr::lag() masks stats::lag()
✖ lubridate::minute() masks wakefield::minute()
✖ lubridate::month() masks wakefield::month()
✖ lubridate::second() masks wakefield::second()
✖ lubridate::year() masks wakefield::year()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsregs <- c( ## regions in the country
"Ashanti",
"Bono",
"Bono East",
"Ahafo",
"Central",
"Eastern",
"Greater Accra",
"Northern",
"Savannah",
"North East",
"Upper East",
"Upper West",
"Volta",
"Oti",
"Western",
"Western North"
)Generate the data
dset <- tibble(
reg = character(),
gender = factor(),
age = integer(),
income = numeric()
)
for( r in regs){
n <-
r_sample_integer(1, x = c(20:30)) # the number of records to generate for each region. The higher the range, the more time it takes, be conservative
for (i in 1:n) {
dset <-
dset %>%
add_row(
reg = r,
gender = gender(n) |> as.character(),
age = age(n) |> as.integer(),
income = income(n) |> as.numeric()
)
}
}
## Show a sample of the data
dset |> sample_n(5)# A tibble: 5 × 4
reg gender age income
<chr> <chr> <int> <dbl>
1 Savannah Female 84 69635.
2 Upper East Male 71 40048.
3 Northern Female 83 29449.
4 Greater Accra Female 63 3092.
5 Western North Female 28 40313.Number of records generated: 25.
Business time
(?) is an anonymous function. An anonymous function is ran once in the code
and no more references can be made to the function. It is great when used in iterations
as the function is only used within the iteration and discarded afterwards.
Don't worry, you will get around and love it too.
_?_ can be any valid variable name. I often use x- First of all the data is grouped based on the region and nested. The nest function comes from dplyr
- Then we mutate, remember to mutate either creates a new column or modifies an existing one. In this case, we are creating a new one which we call model
- It will contain the lm output for each column.
xfrom the anonymous function, which contains the nested data is supplied as an argument to the data argument
mod_data <- dset %>%
group_by(reg) %>%
nest() %>%
mutate(
model = map(data, (x) lm( x$income ~ ., data = x))
)
mod_data# A tibble: 16 × 3
# Groups: reg [16]
reg data model
<chr> <list> <list>
1 Ashanti <tibble [529 × 3]> <lm>
2 Bono <tibble [784 × 3]> <lm>
3 Bono East <tibble [841 × 3]> <lm>
4 Ahafo <tibble [484 × 3]> <lm>
5 Central <tibble [400 × 3]> <lm>
6 Eastern <tibble [576 × 3]> <lm>
7 Greater Accra <tibble [625 × 3]> <lm>
8 Northern <tibble [784 × 3]> <lm>
9 Savannah <tibble [841 × 3]> <lm>
10 North East <tibble [400 × 3]> <lm>
11 Upper East <tibble [900 × 3]> <lm>
12 Upper West <tibble [400 × 3]> <lm>
13 Volta <tibble [441 × 3]> <lm>
14 Oti <tibble [900 × 3]> <lm>
15 Western <tibble [484 × 3]> <lm>
16 Western North <tibble [625 × 3]> <lm> Extracting our models
- (m) is an anonymous function. It is executed and discarded. The function is not stored in the global environment and cannot be referenced.
- tbl_stack and tbl_regression are from the gtsummary package.
- tbl_stack puts a tbl_object on top of each other.
- tbl_regression displays a regression model, nicely formatted
- the mod_data$model contains many models.
- The map function loops over the values in the column and produces a list of tbl_regression objects – Which is used by the tbl_stack function
stk <- tbl_stack(map(mod_data$model, (m)
tbl_regression(m, conf.int =T, label =
list(age = "Age",
gender = "Gender")
)),
group_header = mod_data$reg) In all its glory
stk %>%
as_hux_table() # require the _huxtable_ package to be installed| Group | Characteristic | Beta | 95% CI | p-value |
|---|---|---|---|---|
| Ashanti | Gender | |||
| Female | — | — | ||
| Male | -1,454 | -5,982, 3,075 | 0.5 | |
| Age | 8.7 | -99, 116 | 0.9 | |
| Bono | Gender | |||
| Female | — | — | ||
| Male | 569 | -3,380, 4,518 | 0.8 | |
| Age | -1.4 | -96, 93 | >0.9 | |
| Bono East | Gender | |||
| Female | — | — | ||
| Male | 969 | -2,756, 4,695 | 0.6 | |
| Age | 40 | -51, 131 | 0.4 | |
| Ahafo | Gender | |||
| Female | — | — | ||
| Male | 988 | -4,260, 6,237 | 0.7 | |
| Age | -187 | -310, -64 | 0.003 | |
| Central | Gender | |||
| Female | — | — | ||
| Male | -161 | -5,503, 5,180 | >0.9 | |
| Age | 28 | -97, 153 | 0.7 | |
| Eastern | Gender | |||
| Female | — | — | ||
| Male | 2,269 | -2,336, 6,874 | 0.3 | |
| Age | 58 | -54, 170 | 0.3 | |
| Greater Accra | Gender | |||
| Female | — | — | ||
| Male | 196 | -4,723, 5,115 | >0.9 | |
| Age | -30 | -145, 86 | 0.6 | |
| Northern | Gender | |||
| Female | — | — | ||
| Male | 294 | -3,615, 4,203 | 0.9 | |
| Age | 18 | -80, 115 | 0.7 | |
| Savannah | Gender | |||
| Female | — | — | ||
| Male | -547 | -4,425, 3,330 | 0.8 | |
| Age | 2.2 | -89, 93 | >0.9 | |
| North East | Gender | |||
| Female | — | — | ||
| Male | -817 | -6,354, 4,720 | 0.8 | |
| Age | -56 | -196, 84 | 0.4 | |
| Upper East | Gender | |||
| Female | — | — | ||
| Male | 4,391 | 565, 8,217 | 0.025 | |
| Age | 80 | -15, 174 | 0.10 | |
| Upper West | Gender | |||
| Female | — | — | ||
| Male | 188 | -5,543, 5,919 | >0.9 | |
| Age | -47 | -186, 93 | 0.5 | |
| Volta | Gender | |||
| Female | — | — | ||
| Male | -346 | -4,997, 4,305 | 0.9 | |
| Age | -17 | -132, 97 | 0.8 | |
| Oti | Gender | |||
| Female | — | — | ||
| Male | 2,185 | -1,684, 6,055 | 0.3 | |
| Age | -86 | -179, 8.1 | 0.073 | |
| Western | Gender | |||
| Female | — | — | ||
| Male | 3,078 | -1,759, 7,916 | 0.2 | |
| Age | 8.2 | -107, 123 | 0.9 | |
| Western North | Gender | |||
| Female | — | — | ||
| Male | 1,832 | -3,080, 6,744 | 0.5 | |
| Age | -4.4 | -123, 114 | >0.9 | |
| CI = Confidence Interval | ||||



I like it, and it is an awesome title 🙂 @KID